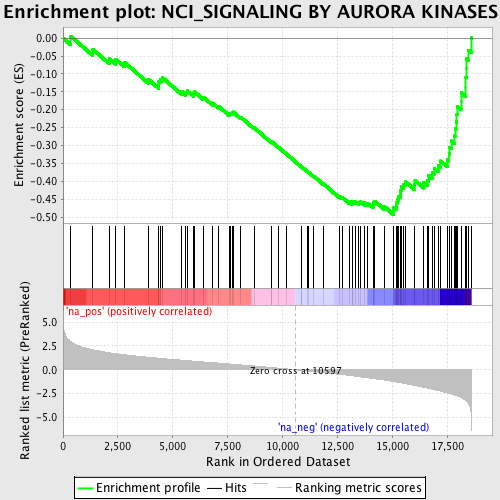
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_wtNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | NCI_SIGNALING BY AURORA KINASES |
| Enrichment Score (ES) | -0.49292612 |
| Normalized Enrichment Score (NES) | -2.1752098 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.004451608 |
| FWER p-Value | 0.017 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ARRB2 | 347 | 2.950 | 0.0056 | No | ||
| 2 | CBX5 | 1358 | 2.087 | -0.0316 | No | ||
| 3 | PPP2R5D | 2112 | 1.784 | -0.0575 | No | ||
| 4 | RELB | 2399 | 1.686 | -0.0590 | No | ||
| 5 | CDC25B | 2787 | 1.580 | -0.0669 | No | ||
| 6 | NFKB2 | 3890 | 1.307 | -0.1156 | No | ||
| 7 | VIM | 4357 | 1.209 | -0.1307 | No | ||
| 8 | PRKACA | 4367 | 1.207 | -0.1212 | No | ||
| 9 | ARHGEF7 | 4422 | 1.194 | -0.1143 | No | ||
| 10 | MYLK | 4541 | 1.166 | -0.1110 | No | ||
| 11 | LCK | 5387 | 1.005 | -0.1483 | No | ||
| 12 | AURKC | 5595 | 0.965 | -0.1515 | No | ||
| 13 | ERC1 | 5649 | 0.957 | -0.1465 | No | ||
| 14 | NFKBIA | 5957 | 0.899 | -0.1556 | No | ||
| 15 | GADD45A | 5991 | 0.894 | -0.1500 | No | ||
| 16 | KIF20A | 6391 | 0.822 | -0.1648 | No | ||
| 17 | SYK | 6811 | 0.741 | -0.1813 | No | ||
| 18 | CDCA8 | 7095 | 0.699 | -0.1908 | No | ||
| 19 | NFKB1 | 7571 | 0.612 | -0.2113 | No | ||
| 20 | DES | 7630 | 0.600 | -0.2095 | No | ||
| 21 | BRCA1 | 7702 | 0.586 | -0.2085 | No | ||
| 22 | OAZ1 | 7752 | 0.575 | -0.2064 | No | ||
| 23 | NPM1 | 8095 | 0.513 | -0.2206 | No | ||
| 24 | PIK3R1 | 8711 | 0.397 | -0.2505 | No | ||
| 25 | TNFAIP3 | 9502 | 0.232 | -0.2912 | No | ||
| 26 | TP53 | 9507 | 0.232 | -0.2895 | No | ||
| 27 | PRKCA | 9829 | 0.161 | -0.3055 | No | ||
| 28 | BUB1 | 10189 | 0.087 | -0.3242 | No | ||
| 29 | FBXW11 | 10879 | -0.066 | -0.3608 | No | ||
| 30 | XPO1 | 10884 | -0.068 | -0.3605 | No | ||
| 31 | PAK1 | 11124 | -0.119 | -0.3724 | No | ||
| 32 | KIF2C | 11196 | -0.138 | -0.3751 | No | ||
| 33 | AKT1 | 11423 | -0.191 | -0.3857 | No | ||
| 34 | RELA | 11860 | -0.290 | -0.4068 | No | ||
| 35 | EVI5 | 12603 | -0.474 | -0.4429 | No | ||
| 36 | SRC | 12725 | -0.502 | -0.4453 | No | ||
| 37 | JUB | 13040 | -0.585 | -0.4574 | No | ||
| 38 | AURKB | 13174 | -0.622 | -0.4595 | No | ||
| 39 | RHOA | 13198 | -0.627 | -0.4555 | No | ||
| 40 | CPEB1 | 13307 | -0.664 | -0.4559 | No | ||
| 41 | RAN | 13479 | -0.712 | -0.4592 | No | ||
| 42 | BCL10 | 13545 | -0.732 | -0.4567 | No | ||
| 43 | STMN1 | 13732 | -0.793 | -0.4602 | No | ||
| 44 | CENPA | 13875 | -0.829 | -0.4610 | No | ||
| 45 | ATM | 14127 | -0.911 | -0.4670 | No | ||
| 46 | BIRC5 | 14134 | -0.913 | -0.4598 | No | ||
| 47 | TNFRSF1A | 14206 | -0.935 | -0.4559 | No | ||
| 48 | PPP1CC | 14643 | -1.074 | -0.4706 | No | ||
| 49 | AURKA | 15058 | -1.229 | -0.4828 | Yes | ||
| 50 | TDRD7 | 15068 | -1.233 | -0.4731 | Yes | ||
| 51 | TACC3 | 15179 | -1.278 | -0.4685 | Yes | ||
| 52 | INCENP | 15209 | -1.289 | -0.4594 | Yes | ||
| 53 | MAP3K14 | 15259 | -1.311 | -0.4512 | Yes | ||
| 54 | TNF | 15287 | -1.321 | -0.4418 | Yes | ||
| 55 | RACGAP1 | 15365 | -1.348 | -0.4348 | Yes | ||
| 56 | GSK3B | 15391 | -1.361 | -0.4249 | Yes | ||
| 57 | KLHL13 | 15435 | -1.378 | -0.4158 | Yes | ||
| 58 | MAPK14 | 15512 | -1.409 | -0.4083 | Yes | ||
| 59 | REL | 15586 | -1.441 | -0.4004 | Yes | ||
| 60 | SMC4 | 15999 | -1.626 | -0.4092 | Yes | ||
| 61 | IKBKB | 16037 | -1.645 | -0.3976 | Yes | ||
| 62 | NOD2 | 16437 | -1.826 | -0.4040 | Yes | ||
| 63 | SGOL1 | 16597 | -1.909 | -0.3969 | Yes | ||
| 64 | BCL3 | 16659 | -1.936 | -0.3842 | Yes | ||
| 65 | PIK3CA | 16832 | -2.033 | -0.3767 | Yes | ||
| 66 | NSUN2 | 16906 | -2.069 | -0.3635 | Yes | ||
| 67 | UBE2D3 | 17090 | -2.167 | -0.3555 | Yes | ||
| 68 | CYLD | 17208 | -2.236 | -0.3433 | Yes | ||
| 69 | KIF23 | 17511 | -2.417 | -0.3397 | Yes | ||
| 70 | NCL | 17587 | -2.462 | -0.3234 | Yes | ||
| 71 | IKBKG | 17625 | -2.489 | -0.3049 | Yes | ||
| 72 | MALT1 | 17690 | -2.531 | -0.2874 | Yes | ||
| 73 | TRAF6 | 17826 | -2.620 | -0.2731 | Yes | ||
| 74 | PSMA3 | 17865 | -2.659 | -0.2532 | Yes | ||
| 75 | CUL3 | 17909 | -2.691 | -0.2333 | Yes | ||
| 76 | NDEL1 | 17946 | -2.716 | -0.2128 | Yes | ||
| 77 | CHUK | 17977 | -2.746 | -0.1917 | Yes | ||
| 78 | RASA1 | 18140 | -2.908 | -0.1764 | Yes | ||
| 79 | TPX2 | 18142 | -2.908 | -0.1525 | Yes | ||
| 80 | NCAPH | 18356 | -3.228 | -0.1373 | Yes | ||
| 81 | MDM2 | 18357 | -3.228 | -0.1107 | Yes | ||
| 82 | BIRC2 | 18371 | -3.248 | -0.0846 | Yes | ||
| 83 | RIPK2 | 18373 | -3.252 | -0.0578 | Yes | ||
| 84 | NDC80 | 18478 | -3.521 | -0.0343 | Yes | ||
| 85 | SMC2 | 18605 | -5.050 | 0.0006 | Yes |
